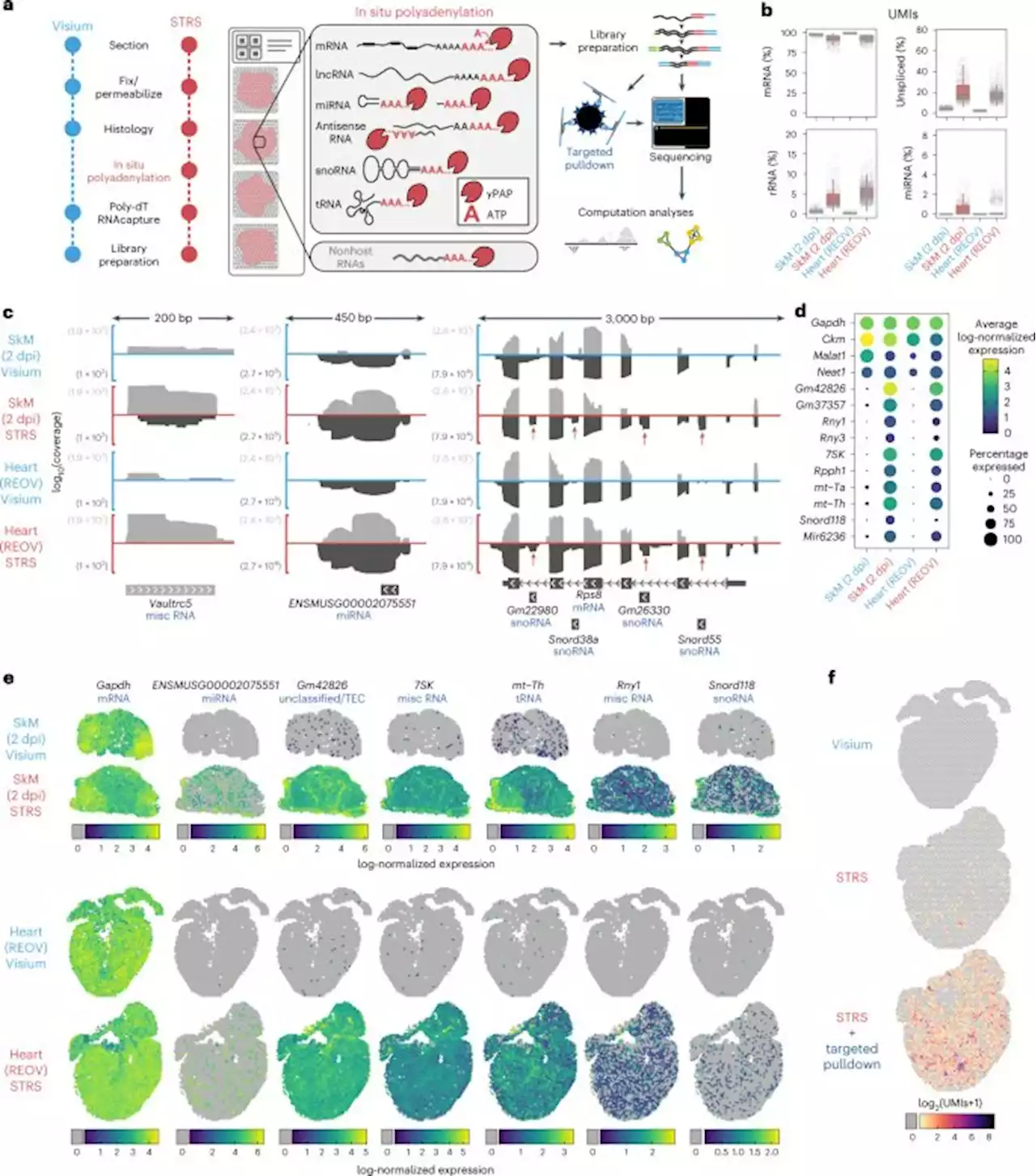
Spatial mapping of the total transcriptome by in situ polyadenylation
. Furthermore, the Visium Gene Expression protocol uses a tagmentation-based library preparation that depletes short molecules by either cutting the UMI/spot barcode or by producing a read that is too short to confidently align to the genome. Despite these issues, we showed robust detection for several known myomiRs and strong correlation with a gold-standard bulk method that does not suffer from ligation or length biases.
This work highlights opportunities for improvements in current bioinformatic tools and resources for single-cell and spatial transcriptomics. Current alignment and transcript counting tools are not optimized for total RNA data and genome annotations are incomplete outside of protein coding genes. Furthermore, new tools that go beyond UMI counts and better leverage the wealth of information in sequence read alignment patterns are likely to be impactful.
On the day of the experiment, nuclei were removed from −20 °C and incubated on ice for 5 min. Nuclei were then pelleted by centrifugation at 1,000O). Nuclei were then pelleted by centrifugation at 1,000, at 4 °C, for 5 min and washed in 200 µl 1× wash buffer . In situ polyadenylation was then performed by suspending nuclei in 50 µl yPAP enzyme mix and incubating at 37 °C for 25 min without agitation.
Small RNA sequencing was performed at the Genome Sequencing Facility of Greehey Children’s Cancer Research Institute at the University of Texas Health Science Center at San Antonio. Libraries were prepared using the TriLink CleanTag Small RNA Ligation kit . Libraries were sequenced with single-end 50× using a HiSeq2500 .
Preprocessing and alignment of STRS, single-nucleus total RNA-sequencing, Smart-Seq-Total and VASA-seq data
Indonesia Berita Terbaru, Indonesia Berita utama
Similar News:Anda juga dapat membaca berita serupa dengan ini yang kami kumpulkan dari sumber berita lain.
 The Coronavirus Standards Working Group’s roadmap for improved population testing - Nature BiotechnologyNature Biotechnology - The Coronavirus Standards Working Group’s roadmap for improved population testing
The Coronavirus Standards Working Group’s roadmap for improved population testing - Nature BiotechnologyNature Biotechnology - The Coronavirus Standards Working Group’s roadmap for improved population testing
Baca lebih lajut »
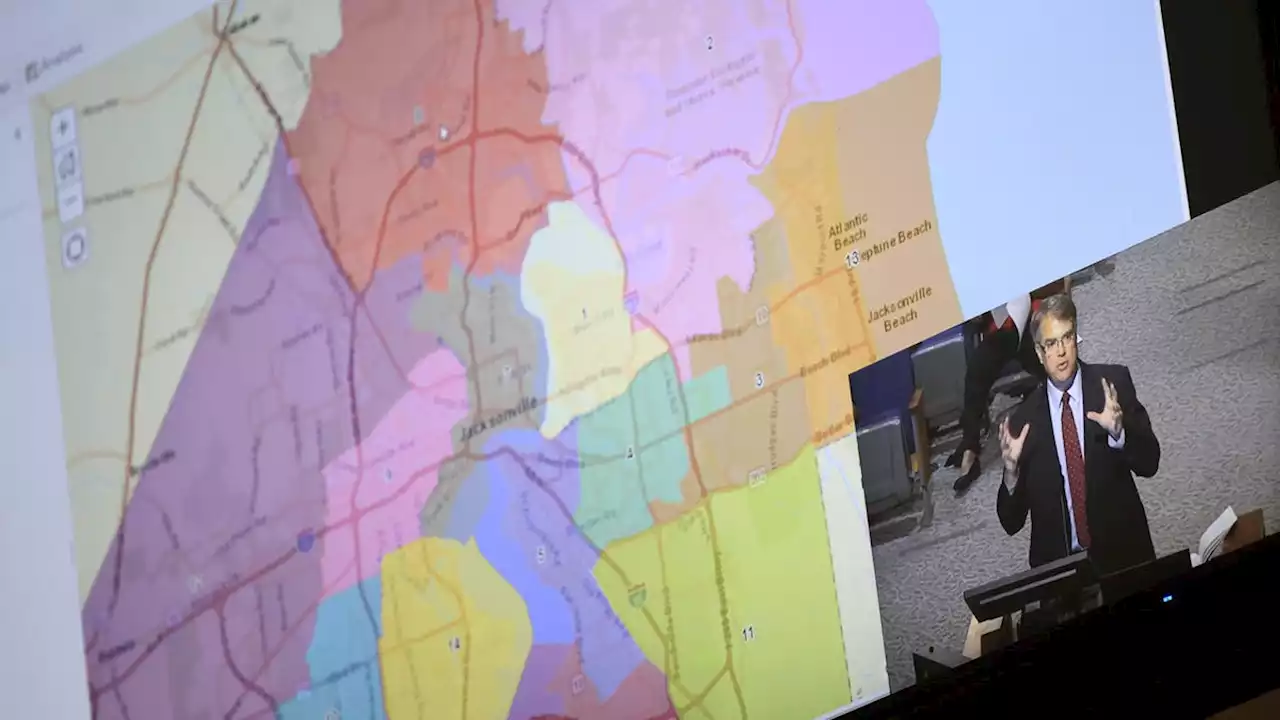 Jacksonville redistricting committee moves forward with 2 maps, removes race as a factorThe redistricting committee disregarded two maps, including the unity map presented by the plaintiffs suing over racially gerrymandered districts.
Jacksonville redistricting committee moves forward with 2 maps, removes race as a factorThe redistricting committee disregarded two maps, including the unity map presented by the plaintiffs suing over racially gerrymandered districts.
Baca lebih lajut »
 Mail-order abortion pill requests surged after Roe reversal, study findsStates with total or near-total bans on abortion saw the largest increases in requests.
Mail-order abortion pill requests surged after Roe reversal, study findsStates with total or near-total bans on abortion saw the largest increases in requests.
Baca lebih lajut »
 A Perfectionist on the BeatDiscipline is second nature to mightbehawa. melindafakuade profiles the composer turned rapper as she releases her debut album, 'Hadja Bangoura'
A Perfectionist on the BeatDiscipline is second nature to mightbehawa. melindafakuade profiles the composer turned rapper as she releases her debut album, 'Hadja Bangoura'
Baca lebih lajut »